during these last months I have been working on applying all the knowledge that I acquired during my secondments in SUH and ROCHE, on building the CLARIFY TNBC Database with Umay Kiraz (ESR11) and researching particularly in the mitosis detection task and the prediction of the Molecular Subtype of the tumor.
In my last Blog Post, I wrote about my experience during my Secondment in the Stavanger University Hospital (SUH) in Stavanger, Norway, where I expanded my knowledge of pathology in breast cancer. However, today, I will update you with advances in my project since I came back to the UPV.
I have been working in parallel on two research lines related to Breast Cancer. The first one, which I have already introduced in my previous posts, is related to the Mitoses Detection in Whole-Slide Images (WSIs). The second one is related with the prediction of global labels, related with the diagnosis and the prognosis in WSIs using a public database that includes biopsies images and the clinical information of the patients.
Regarding the Mitoses Detection task, we were facing a very big problem related with the high number of False Positives in the predictions of our model due to the difficulty of the task. In the mitotic process, different stages correspond to different morphologies. Also, there exist typical and atypical mitoses which also correspond to changes in the morphology of the mitotic figures. All these particularities contribute to pose a hard task to tackle from the Computer Vision perspective (Figure 1).
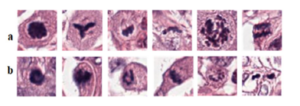
Figure 1. Histopathology patches from TUPAC16 dataset showing heterogeneity in mitosis appearance and their close similarity with non-mitosis, panels (a, b) showing mitotic and non-mitotic nuclei, respectively.
After having our first attempts using an image segmentation using a UNet (Figure 2), we changed our strategy towards binary image classification task, where we used GoogleNet (Figure 3) to classify the patches on whether they contained mitoses or not using the public database TUPAC16. The results of the GoogleNet were also quite poor in terms of False Positives (Figure 4). However, we are trying to overcome this problem by taking advantage of the type of data that we have available in the TUPAC16 Dataset, where the coordinates of the position of the mitotic figures in the images are also provided. Therefore, our aim is to add an additional term to the loss function (Binary Cross-Entropy Loss) which is the Log-Barrier Loss based on the distance between the weighted centroid from the Class Activation Maps (CAMs) of the patches from the class “Mitosis” and the actual centroid provided by our dataset. Finally, we decided to experiment using different convolutional networks architectures such as ResNet or EfficientNet. Good news is that the first experiments with this approach are promising.
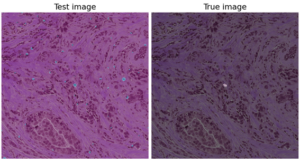
Figure 2. Example of the results from the Mitoses Segmentation model using a UNet which include a high number of false positive mitoses.
Our second research line is related to the prediction of global labels. This corresponds to a weakly-supervised task, and it is approached using Multiple Instance Learning (MIL) techniques. Let me explain, why this is a weakly-supervised problem. Let’s say that we have a Breast Cancer WSI, and we know that it corresponds to a tumor of Grade 3. However, as there are different tissue structures in the image, we don’t know which patches correspond to this classification, but we can say that a “bag” of several patches corresponds to Grade 3, and using (MIL), given a “bag” of patches, our model will be able to predict whether the whole “bag” corresponds to Grade 3.

Figure 3. Pipeline of the Mitoses Classification model using a GoogleNet.
In the last weeks we have started to experiment with the BCNB dataset, which is a public database which includes the WSIs, an annotated region of interest (ROI) and the clinical information of from each patient. This last item is what we are most interested in, as they provide information regarding tumor type, information about the hormone receptors, tumor proliferation markers (Ki-67), the molecular subtype and information related to the status of the Axillary Lymph Nodes (ALN) and how many of them are affected by metastases. This database is particularly interesting for our research because all these variables are also present in the clinical information of the CLARIFY TNBC Database that we are building together with Umay Kiraz (ESR11). Therefore, we can use our own database to test the trained models on the public dataset.
In this sense, our first approach is to try to predict the Molecular Subtype (Luminal A, Luminal B, HER2+ and TNBC) as the treatment that the patient receives is highly influenced by this diagnostic. Also, our idea is to be able to predict the status of the ALN, which corresponds to loco-regional metastasis. Other variables that will be experimented with in the future are the Ki-67 (High or Low Proliferation) and the Tumor Infiltrating Lymphocytes (TILs).
In conclusion, during these last months I have been working on applying all the knowledge that I acquired during my secondments in SUH and ROCHE, on building the CLARIFY TNBC Database with Umay Kiraz (ESR11) and researching particularly in the mitosis detection task and the prediction of the Molecular Subtype of the tumor.
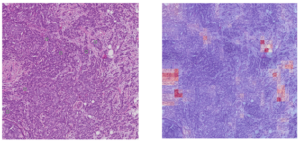
Figure 4. Example of the results form the Mitoses Classification model using the GoogleNet which include a high number of false positive mitoses.
Claudio Fernández – ESR6.

