A comprehensive model takes time and needs step-by-step trials and tests.
Prostate cancer is one of the most common cancers in the male group. Many types of this cancer grow slowly and may need some minimum or no medical treatment. Besides, there are some aggressive types of prostate cancer that can spread quickly and they would be harmful for the patients. If pathologists detect prostate cancer earlier, it can have more chance for successful treatment (the sooner, the better). Therefore, prostate cancer has different grades which need different medication and this variety may make pathologists confused in detecting the grade and selecting the right medical drugs.
According to the rate of men who suffer from prostate cancer, SICAPv2 has been published by UPV to collect a large number of Whole Slide Images (WSIs) of prostate cancer by considering 4 grades which contain Non-Cancerous (NC), Gleason Grade 3 (GG3), Gleason Grade 4 (GG4), Gleason Grade 5 (GG5). Figure1 shows a sample for each of these Gleason grade.
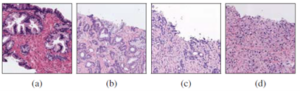 Fig. 1: Examples of histology prostate regions. (a): Noncancerous glands, (b): Gleason pattern 3, (c): Gleason pattern 4 and (d) Gleason pattern 5.
Fig. 1: Examples of histology prostate regions. (a): Noncancerous glands, (b): Gleason pattern 3, (c): Gleason pattern 4 and (d) Gleason pattern 5.
One of the problems that computer aid can cover is grading the new input, which means classifying the new sample among different groups. This can help pathologists to answer a question, wheatear it is aggressive or not? The answer to this question can increase the chance of successful treatment. But how can pathologists rely on its results? They need to see some similar images according to their query to be able to compare them with their knowledge. To do this, Content-Based Image Retrieval (CBIR) brings the outputs which they need.
As we discussed before, to define a CBIR network, a Feature Extractor (FE) is needed to be able to find similarities based on the distances between extracted features. A Convolutional Auto-Encoder (CAE) modified by a residual block plays a role as our FE. Currently, I could finish the pipeline and I fed my network with Mnist, CIFAR-10, and PatchCamelyon (PCAM) dataset. Tests have been started from Mnist since it is the simplest public dataset and it can clarify any problems in the network. As the SICAPv2 is a RGB dataset, I moved to CIFAR-10 to check the capability of the network on RGB images. By having a promising accuracy on these simple datasets, the model has been fed by PCAM as a more complicated dataset. This means I tried to train and test the proposed model step by step with the different datasets to analyze all aspects of it.
As a plan, I am going to feed the network with SICAPv2. By finding an optimum result on this dataset, I will train the proposed model with skin cancer dataset which is preparing by CLARIFY project
Zahra Tabatabei – ESR9.

